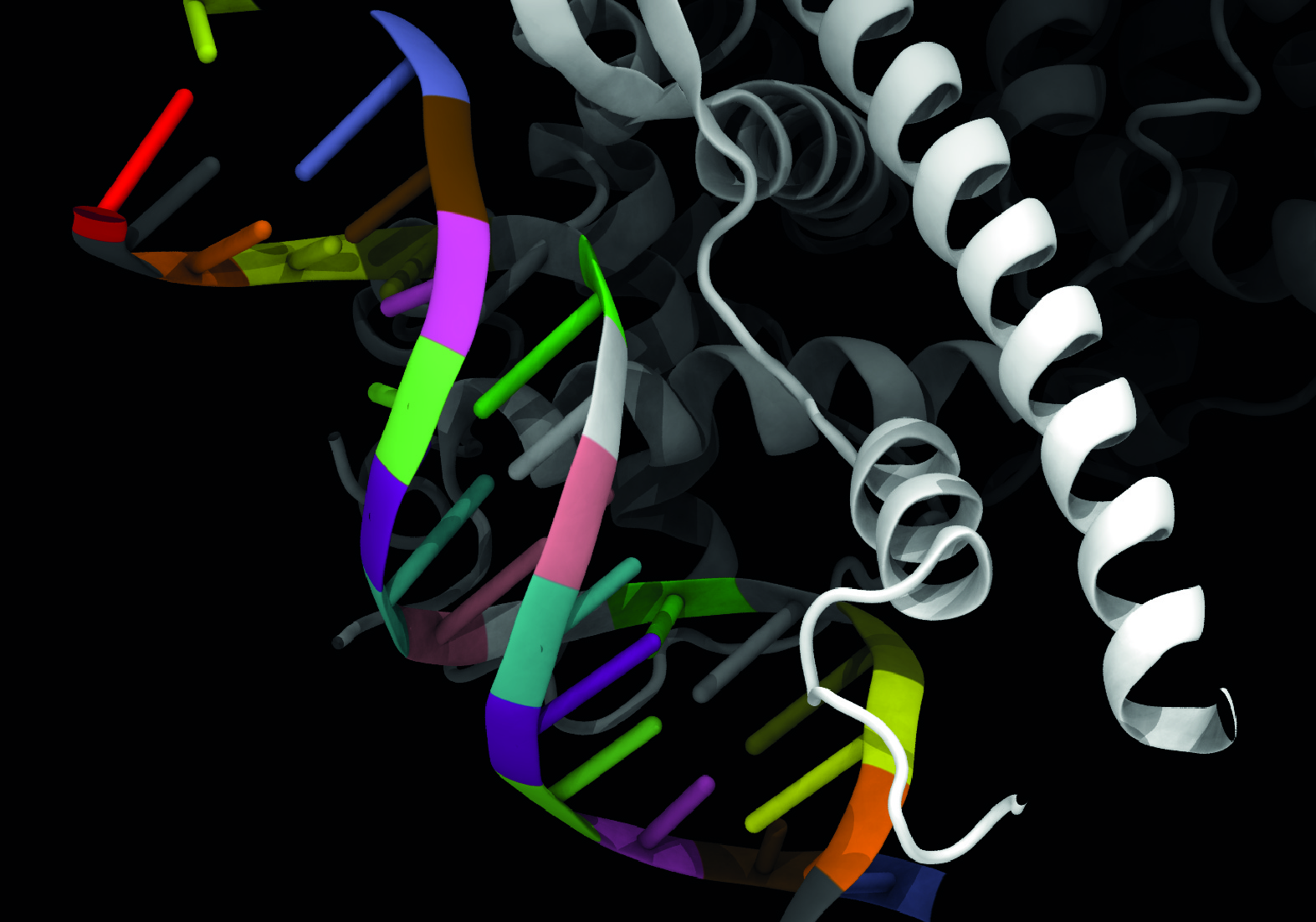
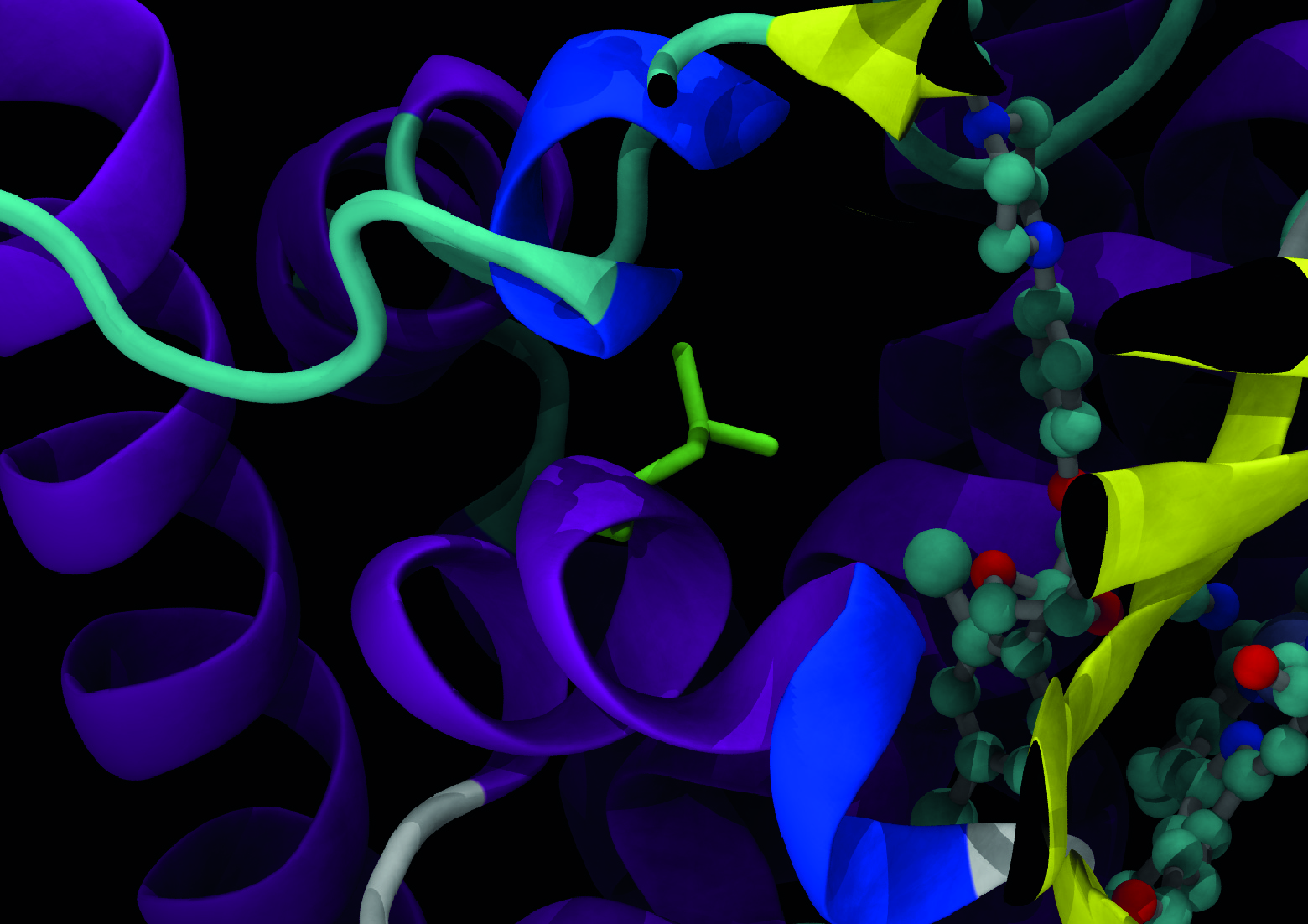
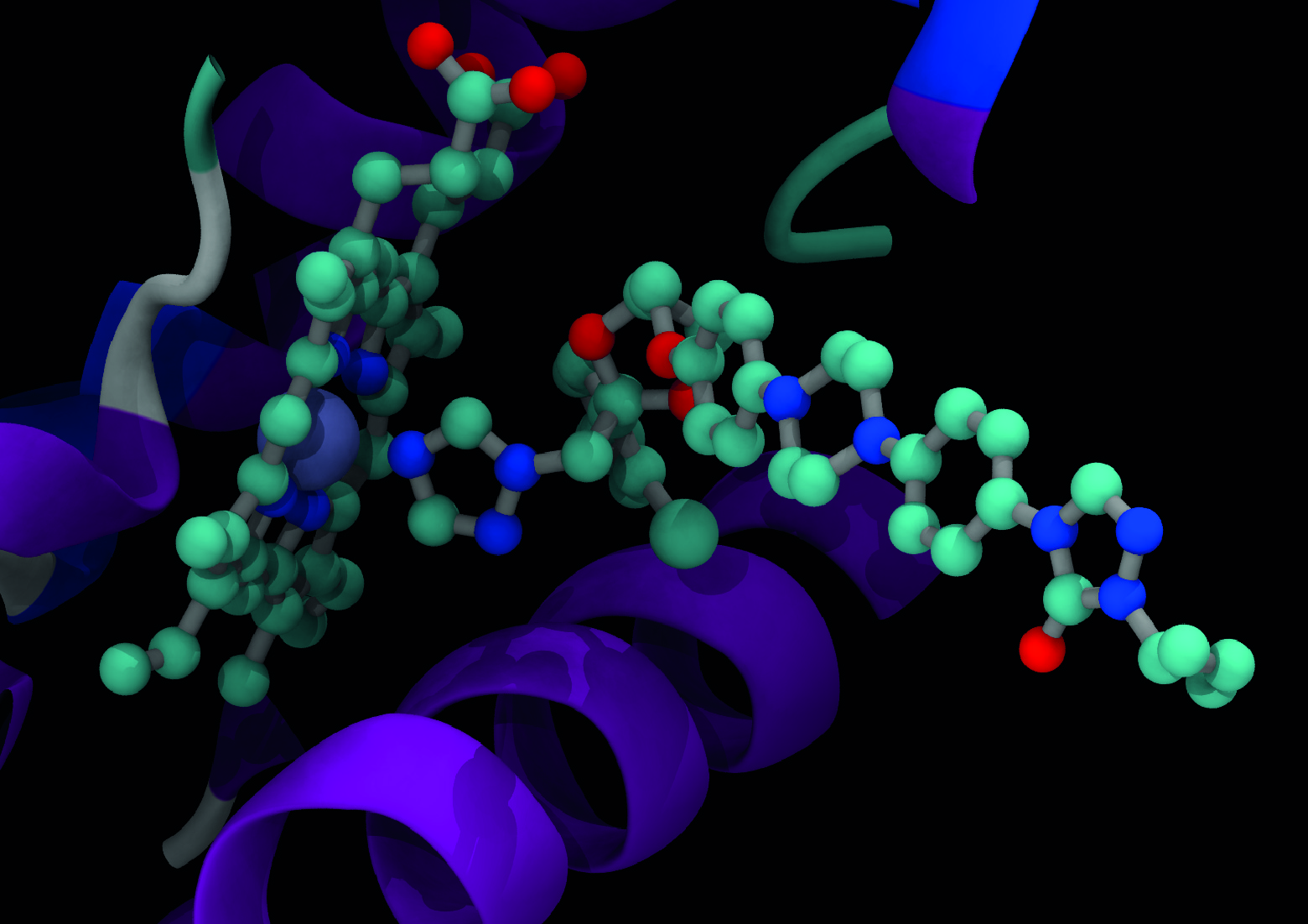
What is MARDy?
MARDy is a curated database of known resistance mechanisms and associated antifungal drugs. There are four major classes of drugs for the treatment of fungal infections: polyenes, azoles, echinocandins, and antimetabolites. Antifungal drug resistance is a complex phenomenon involving multiple mechanisms. MARDy includes resistances as a product of amino acid substitutions, tandem repeat gene sequences, and chromosome ploidy.
MARDy is the intellectual fabrication of Dr Johanna Rhodes of Imperial College London, and it was developed by Dr Anthony Nash of the University of Oxford. The service is hosted and maintained by members of the Fisher Group, directed by Professor Matthew Fisher of Imperial College London. Enquires can be directed through the email link found at the bottom of each page.